

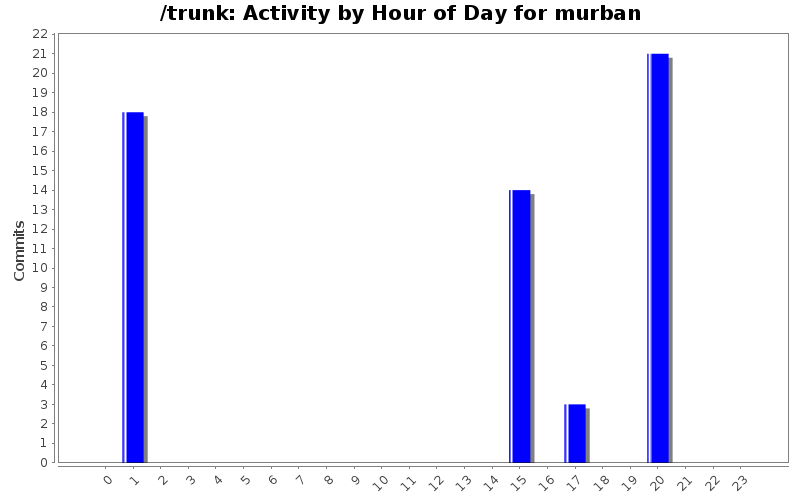
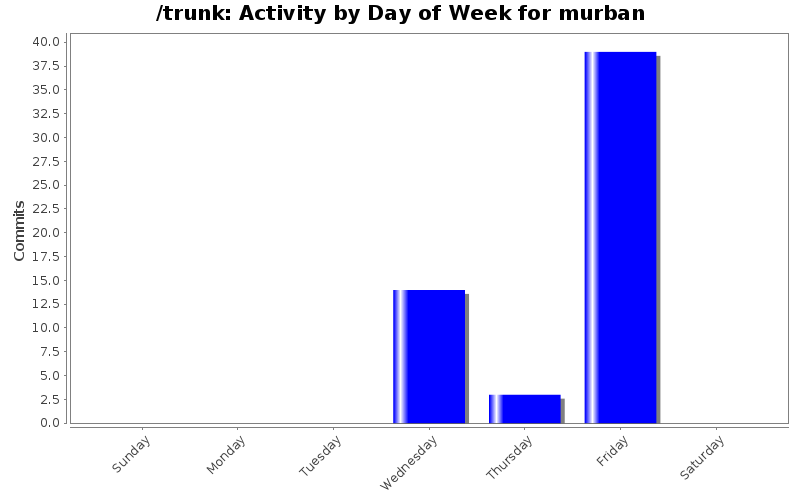
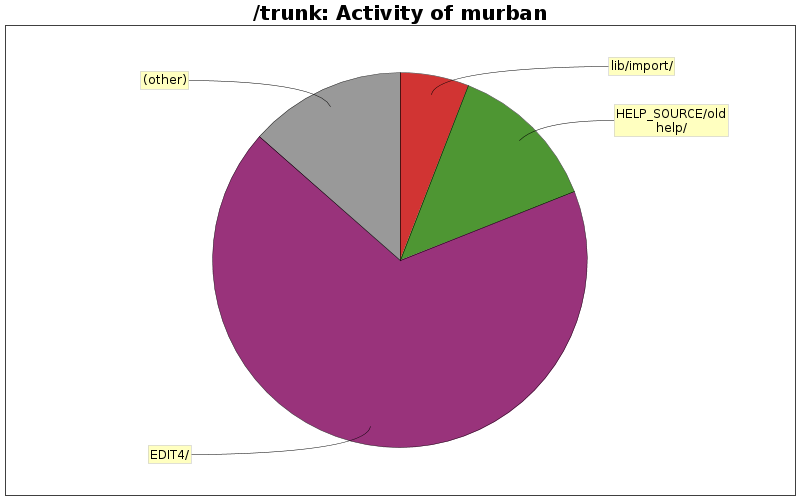
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 56 (100.0%) | 3153 (100.0%) | 56.3 |
| EDIT4/ | 23 (41.1%) | 2127 (67.5%) | 92.4 |
| HELP_SOURCE/oldhelp/ | 10 (17.9%) | 414 (13.1%) | 41.4 |
| lib/import/ | 8 (14.3%) | 187 (5.9%) | 23.3 |
| PERL_SCRIPTS/ARBTOOLS/IFTHELP/ | 3 (5.4%) | 141 (4.5%) | 47.0 |
| NTREE/ | 5 (8.9%) | 123 (3.9%) | 24.6 |
| PERL_SCRIPTS/IFTHELP/ | 2 (3.6%) | 108 (3.4%) | 54.0 |
| lib/protein_2nd_structure/ | 5 (8.9%) | 53 (1.7%) | 10.6 |
bugfix: struct pfold_match_type_awars changed
4 lines of code changed in 1 file:
added doxygen comments; added help; bugfix: order of amino_acids
1200 lines of code changed in 17 files:
updated functions for protein structure comparison; added menu entry in EDIT4 for protein match settings; added help
863 lines of code changed in 14 files:
added functions for protein structure comparison; added import filters and perl script for dssp files; added menu entry in EDIT4 for choosing the active protein structure SAI
1042 lines of code changed in 21 files:
- dssp import filter
6 lines of code changed in 1 file:
- dssp -> fasta converter
38 lines of code changed in 2 files: