Lines of Code
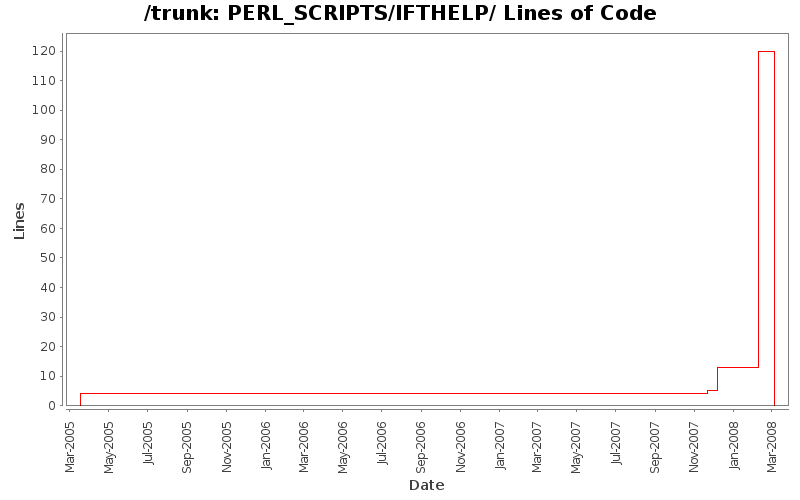
[root]/PERL_SCRIPTS/IFTHELP

| Author | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 12 (100.0%) | 126 (100.0%) | 10.5 |
| murban | 2 (16.7%) | 108 (85.7%) | 54.0 |
| westram | 10 (83.3%) | 18 (14.3%) | 1.8 |
- moved in REPOSITORY to ../ARBTOOLS/IFTHELP
0 lines of code changed in 4 files:
added functions for protein structure comparison; added import filters and perl script for dssp files; added menu entry in EDIT4 for choosing the active protein structure SAI
107 lines of code changed in 1 file:
- reset mode at end of sequence data.
Now files with multiple entries work as well
4 lines of code changed in 1 file:
- converts secondary structure into 2nd species
5 lines of code changed in 1 file:
- dssp -> fasta converter
1 lines of code changed in 1 file:
- unwrap long qualifiers
- prefix with FT and FTx. Makes feature table
compatible with EMBL format
1 lines of code changed in 1 file:
- fixed perl path
1 lines of code changed in 1 file:
- fix for feature names (may also contain non-identifier characters)
4 lines of code changed in 1 file:
- perl script to change embl feature table for more simple
ift-parsing
3 lines of code changed in 1 file: